“REGULATION OF GENE EXPRESSION” – Research Topics
Molecular and cellular mechanisms of the postembryonic plant development
Postembryonic root development
Novel small-molecule effectors of the intracellular membrane trafficking in plant cells
Cell-, tissue- and organ specificity of hormonal signaling and crosstalk
Epigenetic regulation of the gene expression
Impact of the epigenetic DNA modifications on plant development and abiotic stress response
Dissection of the gene functions in heterologous systems
Structural organization of the plant genome in abiotic stress conditions
Genotoxic assessment of the abiotic stress impact on the plant genome
Molecular and phenotypic characterization of the abiotic stress effect on plants
Molecular and phenotypic markers for analysis of the genetic determinants of plant resistance to abiotic stress
Changes in the levels and activity of key stress-related plant proteins. Proteomic analysis.
Molecular and cellular mechanisms of the postembryonic plant development
Postembryonic root development in model plant species
We investigate genes and signaling pathways involved in root growth and branching using the model plant Arabidopsis thaliana and crops, such as wheat, camelina, etc. Particular attention is given to identifying early regulators that control specification of lateral root founder cells and subsequent highly structured asymmetric and symmetric cell divisions, leading to the formation of lateral root primordia and root branching. We also examine the coordinated auxin-dependent nuclear migration, which is an important prerequisite for asymmetric divisions of the founder cells. Since all stages of root growth and development are largely regulated by the phytohormone auxin, the study of key components of the auxin signaling machinery greatly contributes to understanding the adaptation changes during plant development and in response to adverse environment.
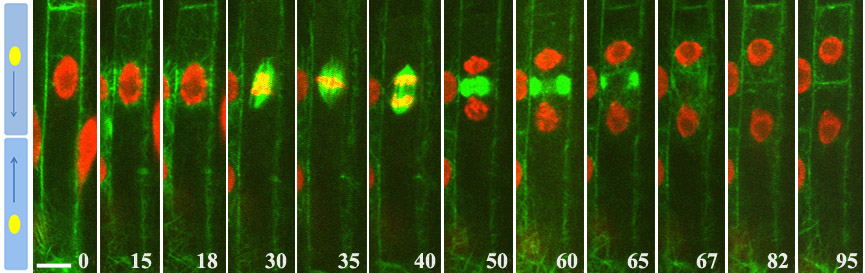
Polarization and asymmetric cell divisions of founder cells in the root pericycle of Arabidopsis thaliana.
Read more
- Velinov V, Zehirov G, Georgieva M, Vassileva V. 2021. NUDC-like genes contribute to root growth and branching in Arabidopsis thaliana. R. Acad. Bulg. Sci. 74, 12, 1767-1773.
- Velinov V, Vaseva I, Zehirov G, Zhiponova M, Georgieva M, Vangheluwe N, Beeckman T, Vassileva V. 2020. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Frontiers in Plant Science, 11, 815.
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, Hilson P. 2015. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. Journal of Experimental Botany 17, 5245-5256.
- Chen Q, Liu Y, Maere S, Lee E, Van Isterdael G, Xie Z, Xuan W, Lucas J, Vassileva V, Kitakura S, Marhavý P. 2015. A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nature Communications 6, 8821.
- Boycheva I, Vassileva V, Revalska M, Zehirov G, Iantcheva A. 2015. Cyclin-like F-box protein plays a role in growth and development of the three model species Medicago truncatula, Lotus japonicus and Arabidopsis thaliana. Research and Reports in Biology 6, 117-130.
- Revalska M, Vassileva V, Zehirov G, Iantcheva I. 2015. Is the auxin influx carrier LAX3 essential for plant growth and development in the model plants Medicago truncatula, Lotus japonicus and Arabidopsis thaliana? Biotechnology & Biotechnological Equipment 29, 4, 786-797.
- Berckmans B, Vassileva V, Schmid S, Maes S, Parizot B, Naramoto S, Magyar Z, Kamei CLA, Koncz C, Persiau G, De Jaeger G, Bogre L, Friml J, Simon R, Beeckman T, De Veylder L. 2011. Auxin-dependent cell cycle reactivation through transcriptional control of E2Fa by LATERAL ORGAN BOUNDARY proteins. Plant Cell 23, 10, 3671-3683.
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Möller B, Holman T, Van Isterdaele G, Brunoud G, Vuylsteke M, Vernoux T, De Veylder L, Inzé D, Weijers D, Bennett M, Beeckman T. 2010. A novel Aux/IAA28 signalling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20, 19, 1697-1706.
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, Wang JY, Meuli N, Vanneste S, Friml J, Hilson P, Jürgens G, Ingram GC, Inzé D, Benfey PN, Beeckman T. 2008. Receptor-like kinase ACR4 restricts stem cell divisions in the Arabidopsis root. Science 322, 5901, 594-597.
[ Top ]
Novel small-molecule effectors of the intracellular membrane trafficking in plant cells
Small bioactive molecules identified in large-scale chemical genetic screens can be helpful in understanding the interplay between endomembrane trafficking and hormone signaling. Chemical genetics overcomes essential limitations of classical forward genetics, such as lethality and genetic redundancy. An important advantage of chemical genetics in dissecting complex biological processes, such as the intracellular trafficking, is the enormous diversity of chemical structures that could be used for probing protein functions in a reversible manner. A major challenge in plant chemical genetics is the identification of protein targets of the bioactive compounds which is essential to understand the mode of action of the small molecules and addresses chemical specificity issues.
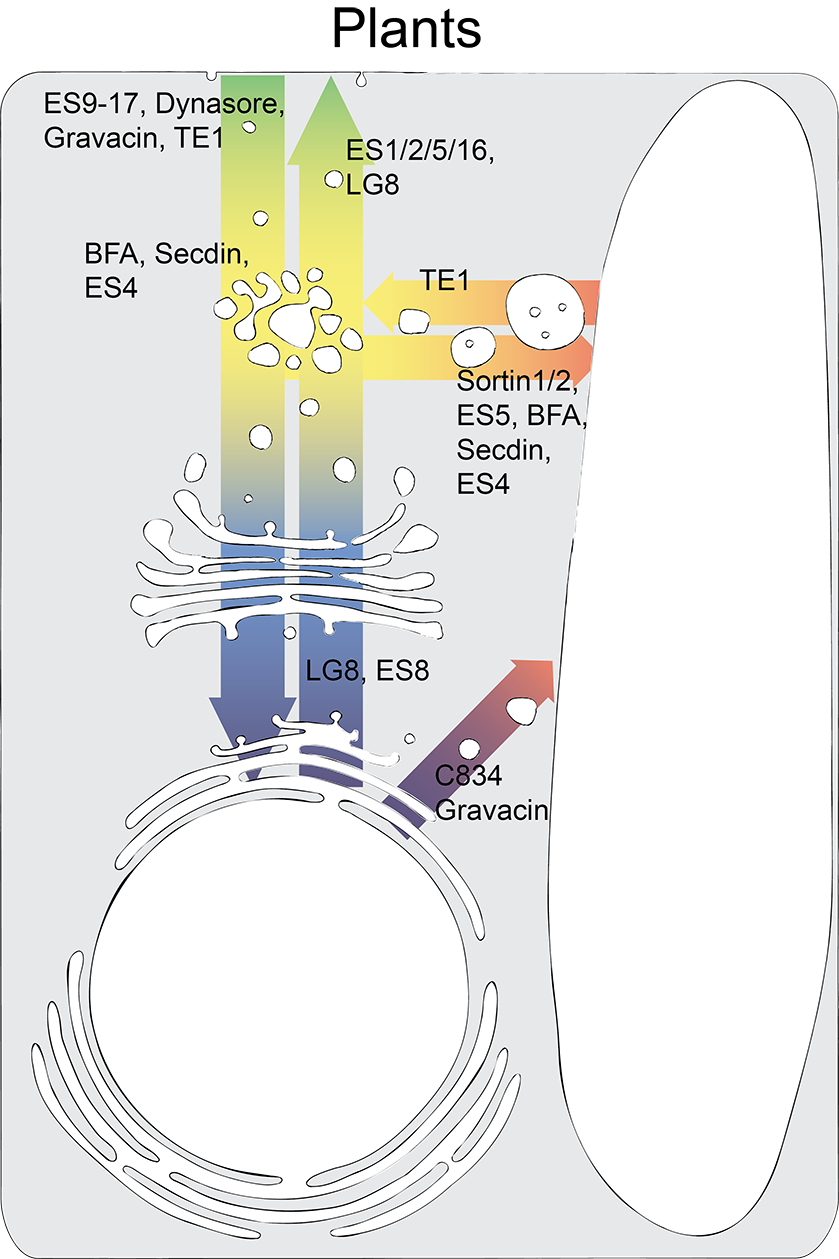
Schematic representation of the main endomembrane trafficking pathways along with the respective small-molecule inhibitors that have been tested so far in plant cells. (Chem Biol 20, 2013, 475-486, updated).
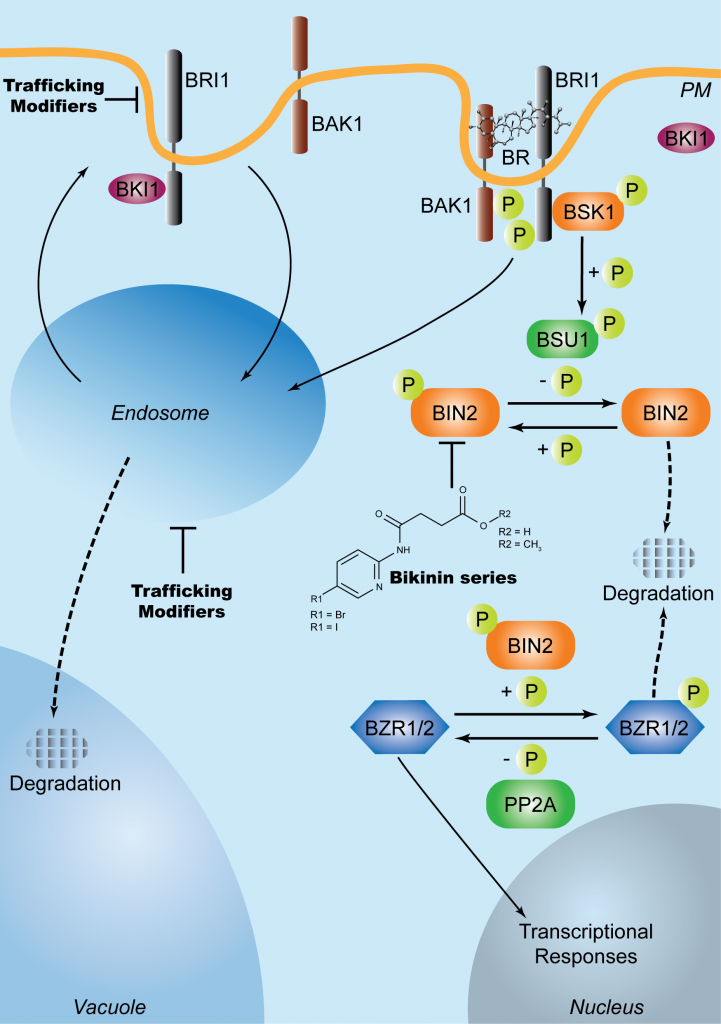
Interplay between the intracellular vesicle trafficking and plant hormone signaling. The direct effect of endocytosis on the hormone signaling outputs has already been demonstrated for the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1), a protein kinase which undergoes constitutive cycling between the plasma membrane and endosomal compartments (Curr Opin Plant Biol 22, 48-55).
Read more
- Dejonghe W, Sharma I, Denoo B, De Munck S, Lu Q, Mishev K, Bulut H, Mylle E, De Rycke R, Vasileva M, Savatin DV, Nerinckx W, Staes A, Drozdzecki A, Audenaert D, Yperman K, Madder A, Friml J, Van Damme D, Gevaert K, Haucke V, Savvides SN, Winne J, Russinova E. 2019. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nature Chemical Biology, 15(6): 641-649.
- Mishev K, Lu Q, Denoo B, Peurois F, Dejonghe W, Hullaert J, De Rycke RM, Boeren S, Bretou M, De Munck S, Sharma IS, Goodman K, Kalinowska K, Storme V, Nguyen L, Drozdzecki A, Martins S, Nerinckx W, Audenaert D, Vert G, Madder A, Otegui MS, Isono E, Savvides S, Annaert W, de Vries SC, Cherfils J, Winne J, Russinova E. Nonselective chemical inhibition of Sec7 domain-containing ARF GTPase exchange factors. The Plant Cell, 30(10): 2573-2593.
- Kania U, Nodzynski T, Lu Q, Hicks GR, Nerinckx W, Mishev K, Peurois F, Cherfils J, De Rycke RM, Grones P, Robert S, Russinova E, Friml J. 2018. The inhibitor Endosidin 4 targets SEC7 domain-type ARF GTPase exchange factors and interferes with subcellular trafficking in eukaryotes. The Plant Cell, 30(10): 2553-2572.
- Dejonghe W, Kuenen S, Mylle E, Vasileva M, Keech O, Viotti C, Swerts J, Fendrych M, Ortiz-Morea FA, Mishev K, Delang S, Scholl S, Zarza X, Heilmann M, Kourelis J, Kasprowicz J, Nguyen le SL, Drozdzecki A, Van Houtte I, Szatmári AM, Majda M, Baisa G, Bednarek SY, Robert S, Audenaert D, Testerink C, Munnik T, Van Damme D, Heilmann I, Schumacher K, Winne J, Friml J, Verstreken P, Russinova E. 2016. Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nature Communications 7, 11710.
- Dejonghe W, Mishev K, Russinova E. The brassinosteroid chemical toolbox. Current Opinion in Plant Biology 22, 48-55.
- Mishev K, Dejonghe W, Russinova E. Small molecules for dissecting endomembrane trafficking: a cross-systems view. Chemistry & Biology 20, 2013, 475-486.
- Irani N, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizoń J, Hniliková J, Šíša M, Buyst D, Buyst D, Vilarrasa-Blasi J, Szatmári AM, Van Damme D, Mishev K, Codreanu MC, Kohout L, Strnad M, Caño-Delgado AI, Friml J, Madder A, Russinova E. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nature Chemical Biology 8, 6, 583-589.
[ Top ]
Cell-, tissue- and organ specificity of hormonal signaling and crosstalk
Cell division and cell expansion that drive growth and development of the organ are controlled by hormones. Hormonal signaling cascades orchestrate a complex network of physiological and biochemical processes as a response to different environmental and developmental cues. The individual cell types have a leading role in the perception of different hormonal signals, which subsequently control various physiological and biochemical processes. Detailed characterization of hormonal network under normal and adverse environmental conditions provides basis for spatiotemporal modelling of hormonal crosstalk allowing examination of distinct and common mechanisms by which hormones control plant growth and stress adaptation. Different hormone signaling mutants, transgenic lines with cell-type specific targeted expression and reporter lines are part of the experimental toolset used in this type of research.
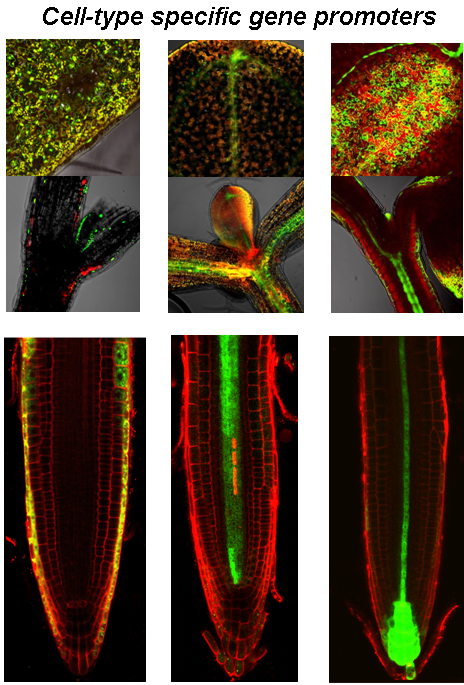
The obtained knowledge could be implemented in development of novel plant breeding strategies or biotechnology tools by precision targeting of growth-regulating processes, which not only can increase stress tolerance but also could be used to improve desired traits in economically important crops.
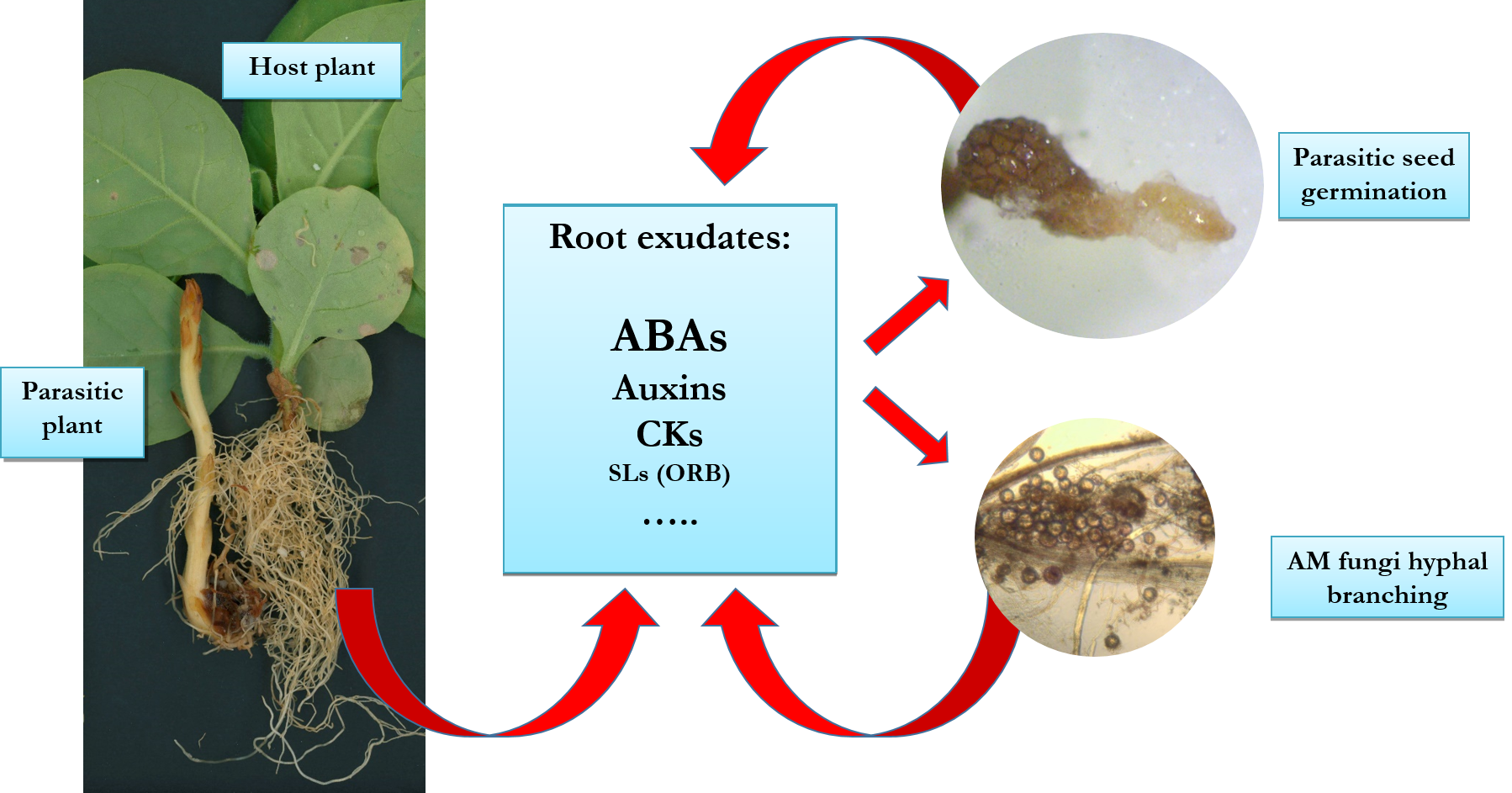
Overview of the belowground interactions of host plants with parasitic plants and arbuscular mycorrhyzal (AM) fungi associated with plant hormone regulation. Co-cultivation of tobacco with broomrape and AM fungi alone or in combination leads to characteristic changes in the levels of endogenous and exuded hormone signals that in turn modulate host plant growth and stress tolerance.
Read more
- Mishev K, Dobrev PI, Lacek J, Filepova R, Yuperlieva-Mateeva B, Kostadinova A, Hristeva T, 2021. Hormonomic changes driving the negative impact of broomrape on plant host interactions with arbuscular mycorrhizal fungi. International Journal of Molecular Sciences, 22(24): 13677.
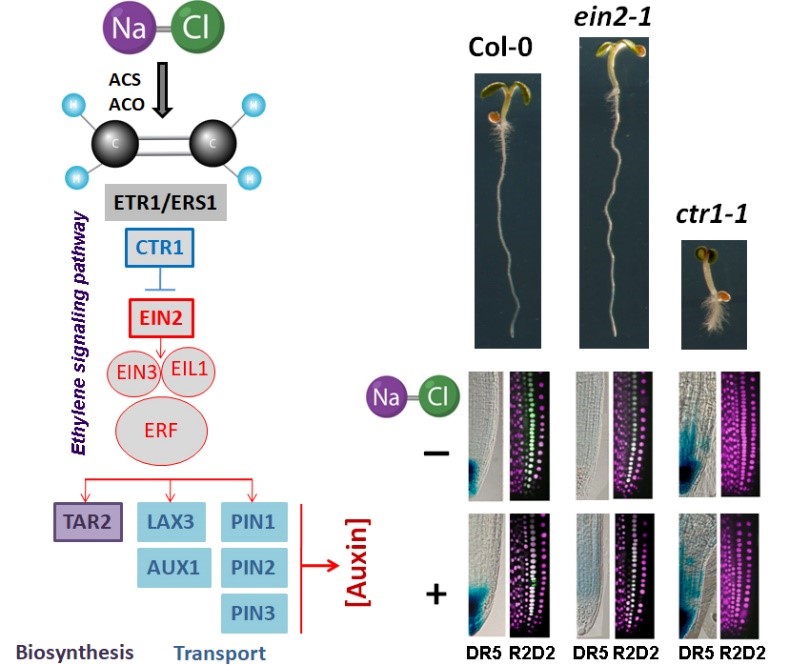
- Vaseva II., Simova-Stoilova L, Kirova E, Mishev K, Depaepe T, Van Der Straeten D, Vassileva V, 2021. Ethylene signaling in salt-stressed Arabidopsis thaliana ein2-1 and ctr1-1 mutants – A dissection of molecular mechanisms involved in acclimation. Plant Physiology and Biochemistry, 167: 999-1010.
- Vaseva II, Mishev K, Depaepe T, Vassileva V, Van Der Straeten D, 2021. The diverse salt-stress response of Arabidopsis ctr1-1 and ein2-1 ethylene signaling mutants is linked to altered root auxin homeostasis. Plants, 10(3): 452.
- Vaseva II, Qudeimat E, Potuschak T, Du Y, Genschik P, Vandenbussche F, Van Der Straeten D. 2018. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proceedings of the National Academy of Sciences of the United States of America, 115 (17): E4130-E4139.
- Zhou J, Liu D, Wang P, Ma X, Lin W, Chen S, Mishev K, Lu D, Kumar R, Vanhoutte I, Meng X, He P, Russinova E, Shan L. 2018. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proceedings of the National Academy of Sciences of the United States of America, 115(8): E1906-1915.
- Vaseva II, Vandenbussche F, Simon D, Vissenberg K, Van Der Straeten D. 2016. Cell type specificity of plant hormonal signals: Case studies and reflections on ethylene. Russian Journal of Plant Physiology, 63, 5, DOI:10.1134/S1021443716050149, 577-586.
- Vandenbussche F, Vaseva I, Vissenberg K, Van Der Straeten D. Ethylene in vegetative development: a tale with a riddle. New Phytologist 194, 4, 895-909.
- Vaseva I, Todorova D, Malbeck J, Trávníčková A, Macháčková I. Response of cytokinin pool and cytokinin oxidase/dehydrogenase activity to abscisic acid exhibits organ specificity in peas. Acta Physiologiae Plantarum 30 (2): 151-155
- Todorova D, Vaseva I, Malbeck J, Trávníčková A, Macháčková I, Karanov E. 2007. Cytokinin oxidase/dehydrogenase activity as a tool in gibberellic acid/cytokinin cross talk. Biol. Plantarum51 (3): 579-583.
- Vaseva-Gemisheva I, Todorova D, Malbeck J, Travnickova A, Machackova I, Karanov E. 2006. Cytokinin pool dynamic changes and distribution of cytokinin oxidase/dehydrogenase activity in peas in relation to developmental senescence. Comp. Rend. Bulg. Acad. Sci. 59 (1): 65-70.
[ Top ]
Epigenetic regulation of the gene expression
Impact of the epigenetic DNA modifications on plant development and stress response
Epigenetic modifications regulate activity of particular genes or non-coding regions of the DNA molecule without changes to the underlying DNA sequence, which provide flexible and reversible regulation of gene expression. In general, epigenetic regulation involves at least three main systems: DNA methylation, histone modification and gene silencing by non-coding RNAs (ncRNA), which control the extent of chromatin condensation. We analyse epigenetically altered regions of the genome, stability of these changes and their inheritance, as well as activity of individual genes. The effects of DNA methylation loss and its potential complementation by direct DNA transfer or genetic crosses are examined, and a correlation between epigenetically-induced changes in gene expression and plant phenotypes are evaluated. The relationship between changed DNA methylation patterns and plant adaptation to abiotic stress has been subject of extensive studies in the recent years. The inherited methylation changes are an important prerequisite for long-term adaptation of plants to changing environments. Part of the research with A. thaliana is dedicated to genes with dense promoter methylation that are involved in stress-related signaling pathways. The methylation status of stress-inducible genes in model plants, as well as in wheat varieties/lines with high tolerance or sensitivity to drought stress, is assessed by means of DNA bisulfite conversion and methylation-sensitive restriction analyses.
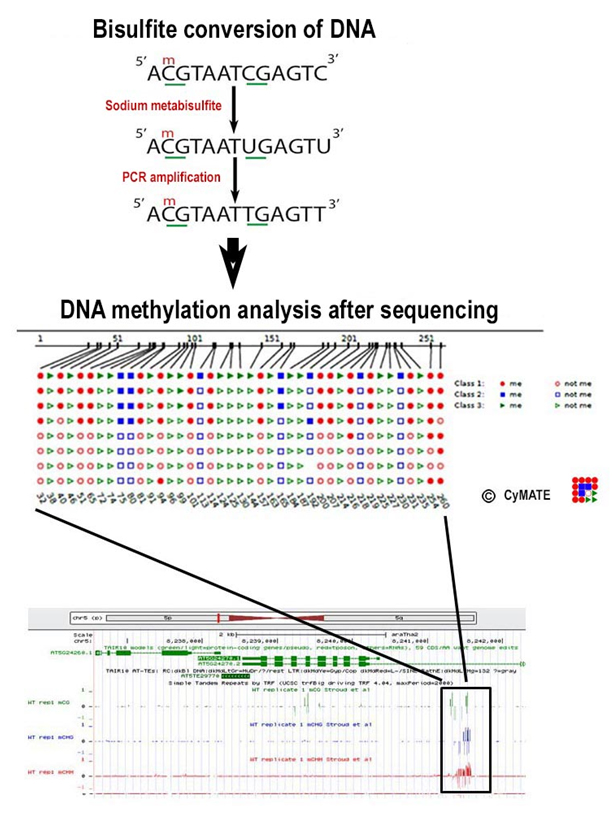
Analysis of the promoter methylation pattern of stress-related genes by bisulfite conversion and subsequent sequencing.
Read more
- Boycheva I, Bonchev G, Manova V, Stoilov L, Vassileva V. How histone acetyltransferases shape plant photomorphogenesis and UV response. International Journal of Molecular Sciences, 25, 14, 7851.
- Agius DR, Kapazoglou A, Avramidou E, Baranek M, Carneros E, Caro E, Castiglione S, Cicatelli A, Radanovic A, Ebejer J-P, Gackowski D, Guarino F, Gulyas A, Hidvegi N, Hoenicka H, Inacio V, Johannes F, Karalija E, Lieberman-Lazarovich M, Martinelli F, Maury S, Mladenov V, Morais-Cecılio L, Pecinka A, Tani E, Testillano PS, Todorov D, Valledor L, Vassileva V. 2023. Exploring the crop epigenome: a comparison of DNA methylation profiling techniques. Front. Plant Sci., 14, 1-25.
- Kakoulidou I, Avramidou EV, Baránek M, Brunel-Muguet S, Farrona S, Johannes F, Kaiserli E, Lieberman-Lazarovich M, Martinelli F, Mladenov V, Testillano PS, Vassileva V, Maury S. 2021. Epigenetics for crop improvement in times of global change. Biology, 10, 8, 766
- Mladenov V, Fotopoulos V, Kaiserli E, Karalija E, Maury S, Baranek M, Segal N, Testillano PS, Vassileva V, Pinto G, Nagel M, Hoenicka H, Miladinović D, Gallusci P, Vergata C, Kapazoglou A, Abraham E, Tani E, Gerakari M, Sarri E, Avramidou Е, Gašparović M, Martinelli F. 2021. Deciphering the epigenetic alphabet involved in transgenerational stress memory in crops. International Journal of Molecular Sciences, 22, 13, 7118
- Boycheva I, Vassileva V, Revalska M, Zehirov G, Iantcheva A. 2017. Role of the histone acetyltransferase HAC1 gene in development of the model species Medicago truncatula, Lotus japonicus and Arabidopsis thaliana. Protoplasma 254, 2, 697-711
- Vassileva V, Hollwey E, Todorov D, Meyer P. 2016. Leaf epidermal profiling as a phenotyping tool for DNA methylation mutants. Genetics and Plant Physiology 6, 1-2, 3-13
- Boycheva I, Vassileva V, Iantcheva A. 2014. Histone acetyltransferases in plant development and plasticity. Current Genomics 15, 1, 28-3
[ Top ]
Dissection of the gene functions in heterologous systems
Model plants are used as alternative experimental systems to study the functions of genes with a high degree of evolutionary conservation. Many human diseases are associated with gene mutations whose lethal effect makes them difficult to study. Plants offer a good alternative, as they are often more tolerant to mutations than animals. Unlike mammals, the model plants, such as A. thaliana, tolerate changes in epigenetic systems, and are viable and fertile when the methylation is lost or reduced. For that reason, we use A. thaliana as an experimental model for studying the degree of conservation and divergence in the specific CG methylation systems. The potential functional replacement of MET1 by DNMT1 that are crucial for methylation maintenance in plants and mammals, respectively, is investigated.
We are also exploring evolutionary conserved genes encoding small heat shock proteins with NudC domain that function not only as molecular chaperones but also play an important role in organismal development. The NudC protein family has representatives in all eukaryotes, including human, which suggest important biological functions preserved during the evolution. We study the NudC genes from A. thaliana, searching functional homology between the plant NudC and yeast (Saccharomyces cerevisiae) genes.

Hypothetical model representing the known and putative functions of NudC genes in different eukaryotes (Plants 2024, 13, 119)
Read more
- Vassileva V, Georgieva M, Todorov D, Mishev K. 2024. Small sized yet powerful: Nuclear Distribution C proteins in plants. Plants, 13, 1, 119.
- Velinov V, Georgieva M, Zehirov G, Vassileva V. 2023. NudC-like genes contribute to root growth and branching in Arabidopsis thaliana. Comptes Rendus de L’Academie Bulgare Des Sciences, 74, 12, 1767-1773.
- Velinov V, Vaseva I, Zehirov G, Zhiponova M, Georgieva M, Vangheluwe N, Beeckman T, Vassileva V. 2020. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Frontiers in Plant Science, 11, 815.
[ Top ]
Structural organization of the plant genome in abiotic stress conditions
Genotoxic assessment of the abiotic stress impact on the plant genome
The genotoxic effects of various environmental abiotic stress factors, such as gamma radiation, UV radiation, chemicals, pesticides, phytocomponents, heavy metals, nanoparticles, etc. are evaluated. The induced plant DNA damage serves as a non-specific biomarker for the environmental impact on plant vigor and is used to predict the effects of various types of pollution in the ecosystems. The analysis of primary DNA damage including single- and double-strand breaks and their repair is performed by means of plant comet assay.
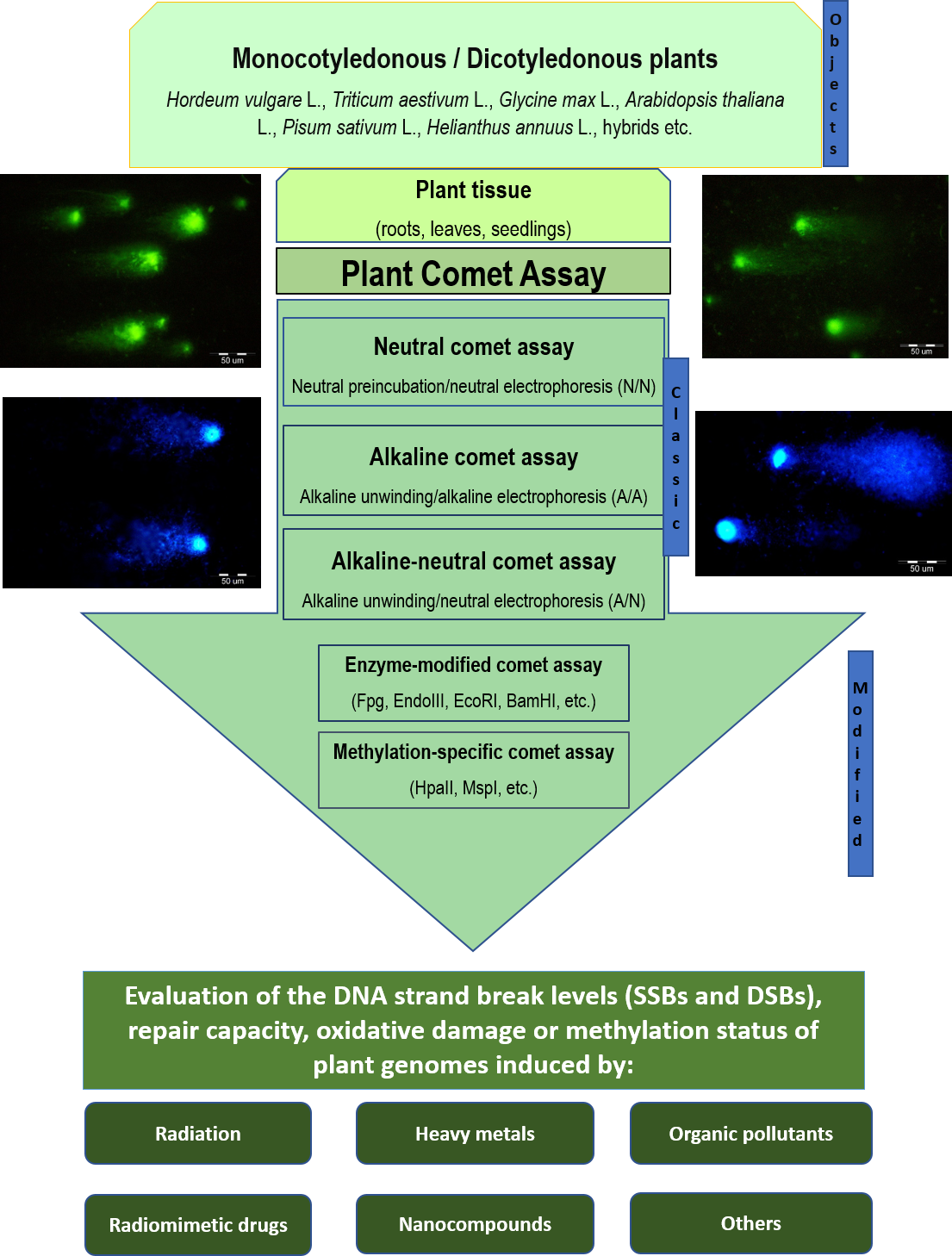
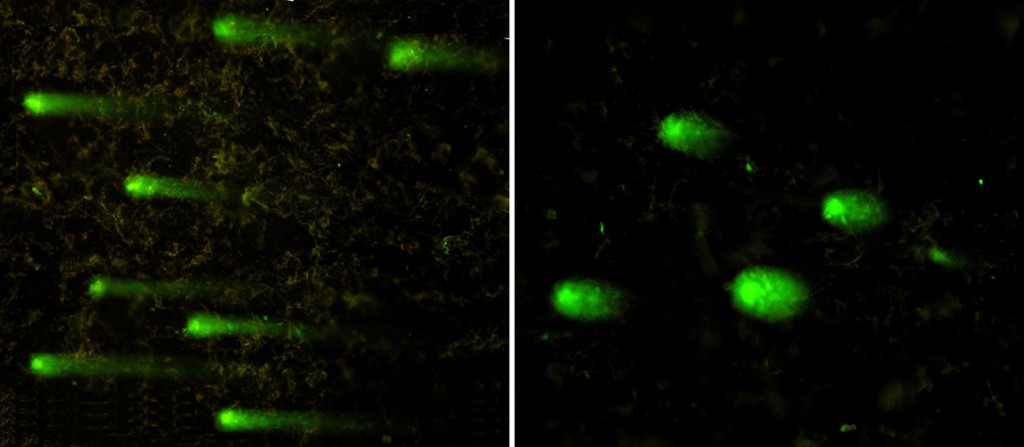
Neutral comet assay on isolated pea nuclei irradiated with UV-C.
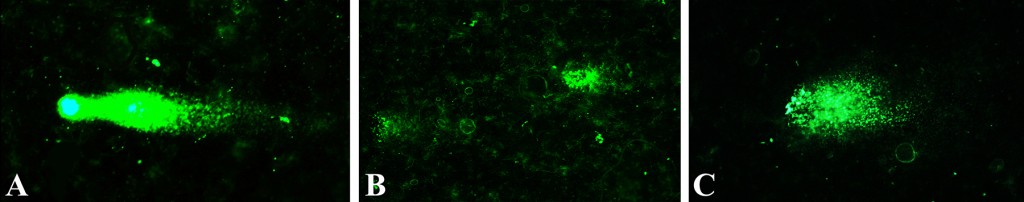
Radionuclide-induced DNA damage (single- and double-strand breaks) in isolated soybean nuclei analyzed through alkaline and alkaline-neutral comet assays.
Read more
- Georgieva M, Bonchev G, Zehirov G, Vasileva V, Vassileva V. 2021. Neonicotinoid insecticides exert diverse cytotoxic and genotoxic effects on cultivated sunflower. Environmental Science and Pollution Research 28, 53193-5320
- Georgieva M, Rashydov NM, Hajduch M. 2017. DNA damage, repair monitoring and epigenetic DNA methylation changes in seedlings of Chernobyl soybeans. DNA repair 50, 14-21.
- Georgieva M, Nikolova I, Bonchev G, Katerova Z, Todorova D. 2015. A comparative analysis of membrane intactness and genome integrity in pea, barley and wheat in response to UVC-irradiation. Turkish Journal of Botany 39, 6, 1008-1013.
- Stoilov L, Georgieva M, Manova V, Liu L, Gecheff K. 2013. Karyotype reconstruction modulates the sensitivity of barley genome to radiation-induced DNA and chromosomal damage. Mutagenesis 28, 2, 153-160.
- Georgieva M, Stoilov L, Rancheva E, Todorovska E, Vassilev D. 2010. Comparative аnalysis of data distribution patterns in plant comet assay. Biotechnology & Biotechnological Equipment 24, 4, 2142-2148.
- Georgieva M, Stoilov 2008. Assessment of DNA strand breaks induced by bleomycin in barley by the comet assay. Environmental and Molecular Mutagenesis 49, 5, 381-387.
[ Top ]
Molecular and phenotypic characterization of the abiotic stress effect on plant
Molecular and phenotypic markers for analysis of the genetic determinants of plant resistance to abiotic stress
We explore the impact of various abiotic stress factors on the survival and productivity of plants, as well as the protective mechanisms by which plants counteract adverse conditions. Our aim is to understand the mechanisms underlying stress adaptability of some genotypes, and to identify traits related to stress tolerance or susceptibility of a particular genotype.
Genotype-dependent physiological, biochemical, molecular, genetic and epigenetic changes in wheat plants under drought stress (Agriculture, 2023, 13, 1823)
We analyze the following physiological, biochemical, molecular, genetic and morphological markers for plant stress tolerance or susceptibility:
- Protein markers: profiles of isoenzymes and storage proteins; the levels of Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase), Rubisco-binding protein, Rubisco activase, heat stress proteins, dehydrins, proteases and enzymes involved in plant cell protection against oxidative stress.
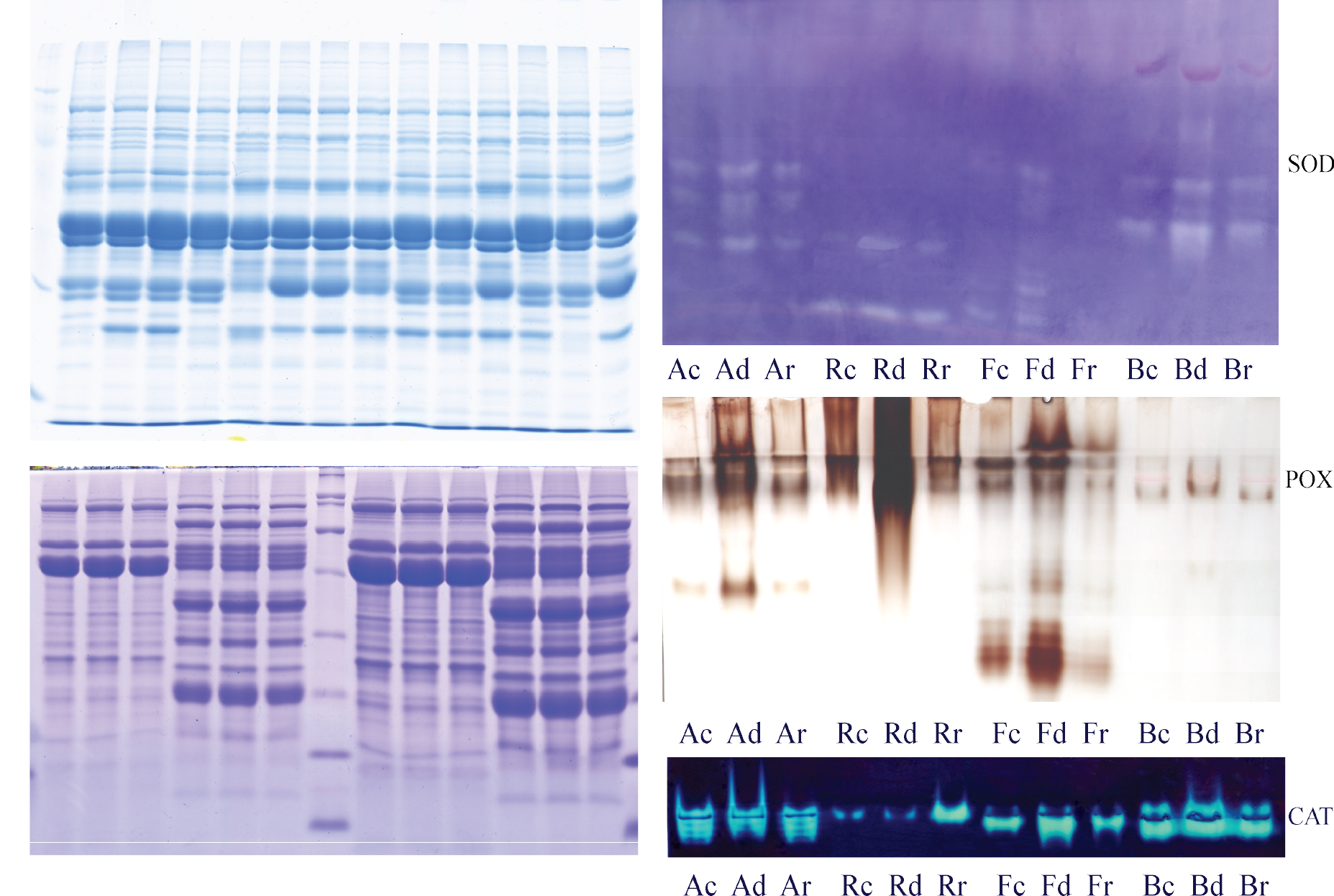
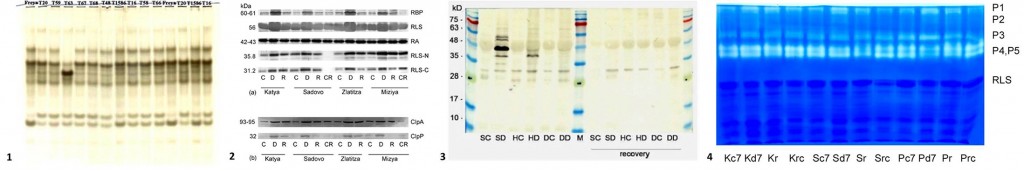
1 – Isoenzyme profile of esterases from mutant barley lines; 2 – Immunoblot analysis of extracts from wheat varieties with different resistance to drought; 3 – Immunoblot analysis of dehydrins in Trifolium; 4 – Protease activity in wheat varieties with different resistance to drought.
- Gene markers: analysis of the expression levels of stress-related genes under different abiotic stress conditions.
- Phenotypic markers: leaf and root phenotyping, plant organ micromorphology under abiotic stress.
Read more
- Georgieva M, Vassileva V. Stress management in plants: examining provisional and unique dose-dependent responses. International Journal of Molecular Sciences, 24, 6, 5105.
- Vassileva V, Georgieva M, Zehirov G, Dimitrova A. 2023. Exploring the genotype-dependent toolbox of wheat under drought stress. Agriculture, 13, 1823.
- Kirova E, Pecheva D, Simova-Stoilova L, 2021. Drought response in winter wheat – protection from oxidative stress and mutagenesis effect. Acta Physiologiae Plantarum 43, 8.
- Simova-Stoilova L, Pecheva D, Kirova E, 2020. Drought stress response in winter wheat varieties – changes in leaf proteins and proteolytic activities. Acta Botanica Croatica 79, 2, 129-138.
- Simova-Stoilova L, Vassileva V, Feller U, 2016. Selection and breeding of suitable crop genotypes for drought and heat periods in a changing climate: which morphological and physiological properties should be considered? Agriculture 6, 26, doi:10.3390/agriculture6020026.
- Vaseva I, Zehirov G, Kirova E, Simova-Stoilova L. 2016. Transcript profiling of serine- and cysteine protease inhibitors in Triticum aestivum varieties with different drought tolerance. Cereal Research Communications 44, 1, 79–88.
- Vaseva I, Anders I, Yuperlieva-Mateeva B, Nenkova R, Kostadinova A, Feller U. 2014. Dehydrin expression as a potential diagnostic tool for cold stress in white clover. Plant Physiology and Biochemistry, 78, 43-48.
- Vaseva I, Anders I, Feller U. 2014. Identification and expression of different dehydrin subclasses involved in the drought response of Trifolium repens. Journal of Plant Physiology, 171, 3-4, 213-224.
- Todorovska E, Dimitrova A, Stoilov L, Christov N, Vassilev D, Gecheff K. 2013. Molecular variability in barley structural mutants produced by gamma-irradiation. Plant Mutation Rep. 3, 1, 4-8.
- Georgieva M, Gecheff K. 2013. Molecular cytogenetic characterization of a new reconstructed barley karyotype. Biotechnology & Biotechnological Equipment 27, 1, 3577-3582.
- Vassileva V, Demirevska K, Simova‐Stoilova L, Petrova T, Tsenov N, Feller U. 2012. Long-term field drought affects leaf protein pattern and chloroplast ultrastructure of winter wheat in a cultivar specific manner. Journal of Agronomy and Crop Science 198, 2, 104–117.
- Grigorova B, Vassileva V, Klimchuk D, Vaseva I, Demirevska K, Feller U. 2012. Drought, high temperature, and their combination affect ultrastructure of chloroplasts and mitochondria in wheat (Triticum aestivum) leaves. Journal of Plant Interactions 7, 3, 204-213.
- Grigorova B, Vaseva I, Demirevska K, Feller U. 2011. Combined drought and heat stress in wheat: changes in some heat shock proteins. Biologia Plantarum 55, 1, 105-111.
- Grigorova B, Vaseva I, Demirevska K, Feller U. 2011. Expression of selected heat shock proteins after individually applied and combined drought and heat stress. Acta Physiologiae Plantarum 33, 5, 2041-2049.
- Vassileva V, Signarbieux C, Anders I, Feller U. 2011. Genotypic variation in drought stress response and subsequent recovery of wheat (Triticum aestivum ). Journal of Plant Research 124, 1, 147-154.
- Vaseva I, Akiscan Y, Demirevska K, Anders I, Feller U. 2011. Drought stress tolerance of red and white clover–comparative analysis of some chaperonins and dehydrins. Scientia Horticulturae, 130, 3, 653-659.
- Vaseva I, Sabotič J, Šuštar-Vozlič J, Meglič V, Kidrič M, Demirevska K, Simova-Stoilova L. 2011. The response of plants to drought stress: the role of dehydrins, chaperones, proteases and protease inhibitors in maintaining cellular protein function. Droughts: New Research, Nova Science Publishers, 1-45
- Simova-Stoilova L, Vaseva I, Grigorova B, Demirevska K, Feller U. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiology and Biochemistry 48, 2-3, 200-206.
- Vaseva I, Grigorova B, Simova-Stoilova L, Demirevska K, Feller U. 2010. Abscisic acid and late embryogenesis abundant protein profile changes in winter wheat under progressive drought stress. Plant Biology, 12, 5, 698-707.
- Vassileva V, Simova-Stoilova L, Demirevska K, Feller U. 2009. Variety-specific response of wheat (Triticum aestivum) leaf mitochondria to drought stress. Journal of Plant Research 122, 4, 445-454.
- Demirevska K, Zasheva D, Dimitrov R, Simova-Stoilova L, Stamenova M, Feller U. 2009. Drought stress effects on Rubisco in wheat: changes in the Rubisco larges ubunit. Acta Physiol. Plantarum 31, 1129-1138.
- Dimitrova А, Todorovska E, Christov NK, Stoilov L, Atanassov A, Gecheff K. 2008. Molecular characterization of structural barley mutants produced by gamma-irradiation. Genetics and Breeding, 37, 3-4, 15-26.
- Demirevska K, Simova-Stoilova L, Vassileva V, Vaseva I, Grigorova B, Feller U. 2008. Drought induced leaf protein alterations in sensitive and tolerant wheat varieties. General and Applied Plant Physiology, Special Issue 34, 1-2, 79-102.
- Demirevska K, Simova-Stoilova L, Vassileva V, Feller U. 2008. Drought response of Rubisco and some ATP-dependent chaperone proteins in wheat plants. Plant Growth Regulation 56, 97–106.
- Kirova E, Tzvetkova N, Vaseva I, Ignatov G. 2008. Photosynthetic responses of nitrate-fed and nitrogen-fixing soybeans to progressive water stress. Journal of Plant Nutrition 31, 445–458.
- Stoilova T, Dimitrova A, Angelova T, Gecheff K. 2006. Assessment of the genetic diversity in barley mutant lines using biochemical markers. Genetics and Breeding 35, 3-4, 3-10.
[ Top ]
Changes in the levels and activity of key stress-related plant proteins. Proteomic analysis.
Proteins are direct effectors in all processes related to cell structure and function, and are essential determinants of the adaptation to the changing environment, underlying the so called phenotypic plasticity. Cell protein composition is highly dynamic and does not necessarily correspond to the transcript levels, due to the complex regulation of gene expression (at genetic, transcriptional, translational, and post-translational levels). The development of databases with partially or fully sequenced plant genomes and expressed sequence tags, necessary for correct protein identification, as well as comprehensive proteome maps of major crops, has facilitated the use of proteomic studies in search of suitable protein markers for assisted selection. Two-dimensional electrophoresis combined with mass spectrometry detects relatively more abundant proteins like key metabolic enzymes, thus providing essential information about changes in the main metabolic pathways and biological processes affected by the stress. Moreover, isoforms and posttranslational modifications of a given protein can also be detected. Deeper proteome coverage, especially for less abundant proteins (signaling, transporters, etc), is achieved by second generation shotgun proteomics, both approaches being complementary.
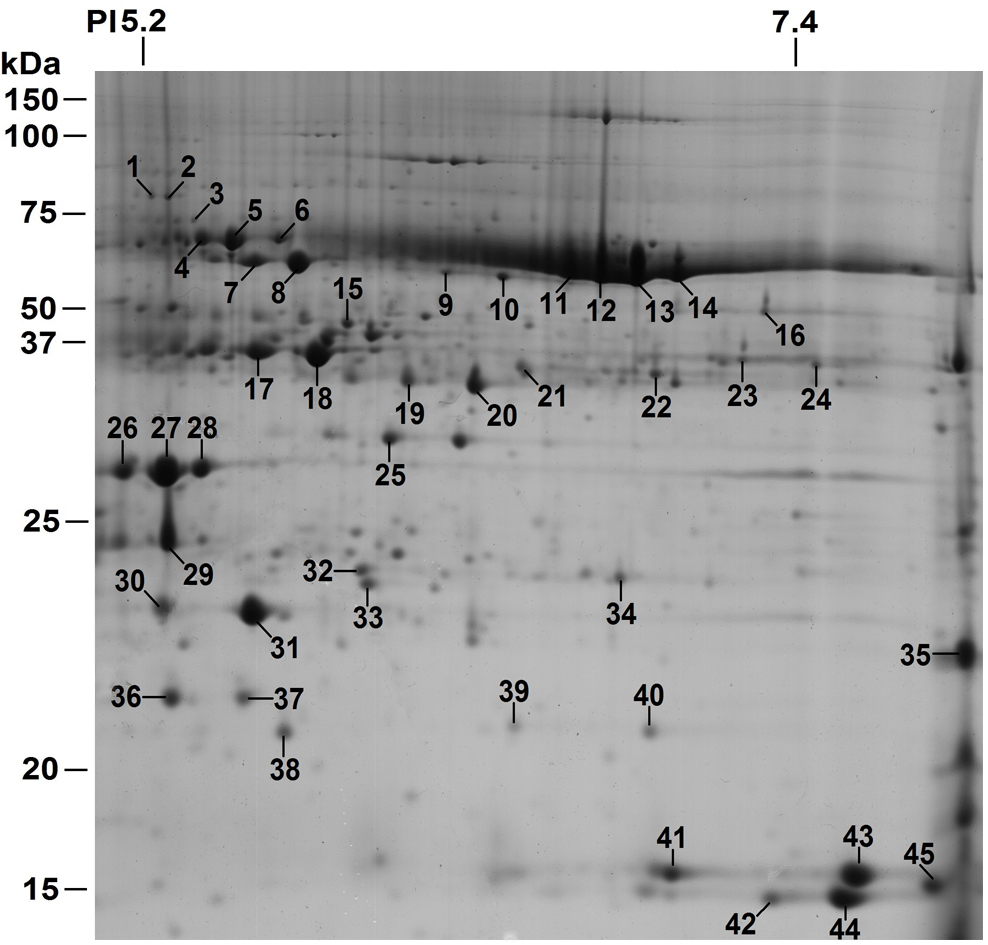
Representative 2DE protein profile of leaf extract from Trifolium. Spots changing in abundance as a result of the applied stress (waterlogging) are indicated by numbers and are identified by MS (Stoychev et al, Advances in Environmental Research, Vol. 39, 2015, 131-162).
Abiotic stresses with a dehydration component (i.e., drought, salt, and freezing) typically trigger an increase in the amount of inactive proteins – denatured, aggregated or oxidatively damaged. The existing protein quality control system maintains proteins in their functional conformation, preventing aggregation of non-native proteins, refolding of denatured proteins to their native conformation, and removing non-functional and potentially harmful polypeptides, which is vital for cell survival under dehydration stress. Plants respond to stress by synthesis of protective proteins, such as dehydrins and chaperones, and degradation of irreversibly damaged proteins by proteases. Protease activity is partially controlled by endogenous protease inhibitors. Plant exposure to stress leads to inhibition of photosynthesis and breakdown of unnecessary and reserve proteins, providing building blocks and energy sources for the metabolism. Some proteins from the quality control system are potential biochemical markers for assessing stress tolerance.
Read more
- Simova-Stoilova LP, López-Hidalgo C, Sanchez-Lucas R, Valero-Galvan J, Romero-Rodríguez C, Jorrin-Novo JV. 2018. Holm oak proteomic response to water limitation at seedling establishment stage reveals specific changes in different plant parts as well as interaction between roots and cotyledons. Plant Science, 276: 1-13.
- Simova-Stoilova L, Vassileva V, Feller U. 2016. Selection and breeding of suitable crop genotypes for drought and heat periods in a changing climate: which morphological and physiological properties should be considered? Agriculture 6, 26, doi:10.3390/agriculture6020026
- Simova-Stoilova LP, Romero-Rodriguez MC, Sánchez-Lucas R, Navarro-Cerrillo RM, Medina- Auñón A, Jorrin-Novo JV. 2015. 2-DE proteomics analysis of drought treated seedlings of Quercus ilex supports a root active strategy for metabolic adaptation in response to water shortage. Frontiers in Plant Science 6, 627.
- Stoychev V, Simova-Stoilova L, Vassileva V, Jorrín Novo JV, Vaseva I, Velikova V, Tsonev T, Demirevska K. 2015. Changes in 2-DE Protein Profile of White and Red Clover Leaves in Response to Waterlogging Stress and Recovery. Advances in Environmental Research 39, 131-162. Ed. Justin A. Daniels, Nova Sci Publ. New York.
- Vaseva II, Anders I, Feller U. 2014. Identification and expression of different dehydrin subclasses involved in the drought response of Trifolium repens. Journal of Plant Physiology 171, 213–224.
- Vaseva II, Anders I, Yuperlieva-Mateeva B, Nenkova R, Kostadinova A, Feller U. 2014. Dehydrin expression as a potential diagnostic tool for cold stress in white clover. Plant Physiology and Biochemistry 78, 43-48.
- Stoychev V, Simova-Stoilova L, Vaseva I, Kostadinova A, Nenkova R, Feller U, Demirevska K. 2013. Protein changes and proteolytic degradation in red and white clover plants subjected to waterlogging. Acta Physiol. Plant. 35, 6, 1925-1932.
- Grigorova B, Vaseva I, Demirevska K, Feller U. 2011. Combied drought and heat stress in wheat: changes in some heat shock proteins. Plantarum 55, 1, 105-111.
- Grigorova B, Vaseva I, Demirevska K, Feller U. 2011. Comparative study of HSP expression after individualy applied and combined drought/heat stress in wheat plants. Acta Physiol. Plant. 33, 2041-2049.
- Vaseva I, Akiscan Y, Demirevska K, Anders I, Feller U. 2011. Drought stress tolerance of red and white clover – comparative analysis of some chaperonins and dehydrins. Scientia Horticulturae 130, 653–659.
- Vaseva I, Sabotič J, Šuštar-Vozlič J, Meglič V, Kidrič M, Demirevska K, Simova-Stoilova L. 2011. Response of plants to drought stress – the role of dehydrins, chaperones, proteases and protease inhibitors in maintaining cellular protein functionIn: Neves DF& Sanz JD (Eds.) Droughts: New Research, Chapter 11, Nova Science Publishers, Inc.
- Simova-Stoilova L, Vaseva I, Grigorova B, Demirevska K, FellerU. 2010. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Phys. Biochem. 48, 2-3, 200-206.
- Vaseva I, Grigorova B, Simova-Stoilova L, Demirevska K, Feller U. 2010. Abscisic acid and late embryogenesis abundant protein profile changes in winter wheat under progressive drought stress. Plant Biology 12, 5, 698-707.
- Simova-Stoilova L, Demirevska K, Petrova T, Tsenov N, Feller U. 2009. Antioxidative protection and proteolytic activity in tolerant and sensitive wheat (Triticum aestivum) varieties subjected to long-term field drought. Plant Growth Regulation 58, 1, 107-117.
