“GENOME DYNAMICS AND STABILITY” – Epigenetics
The investigations are focused on the epigenetic regulation of gene expression addressing the following aspects: 1. Epigenetic mechanisms during the growth and development of eukaryotic organisms – the processes of differentiation, de novo genome methylation, genome imprinting etc.; 2. Epigenetic changes, notably in DNA methylation, as a result of daily use of substances such as caffeine, nicotine and alcohol, substances applied in the therapeutic practice like hormones; and psychological stress as a consequence of traumatic events during person’s lifetime history; 3. Promoter methylation and expression of target genes implicated in human diseases including addictions to psychoactive substances and cancer. Animal model systems and human subjects are studied under the regulation of a local ethics commission.
Epigenetic changes are “changes in gene function that are heritable and that do not entail a change in DNA sequence” (Wu and Morris, 2001). These are modifications in the DNA or the associated proteins, which modulate gene expression by causing changes in chromatin structure thus affecting gene accessibility (Fig. 1). Three major groups of mechanisms for epigenetic regulation could be distinguished that can initiate and sustain epigenetic changes: methylation of DNA, post-translational histone modifications and non-coding RNAs.
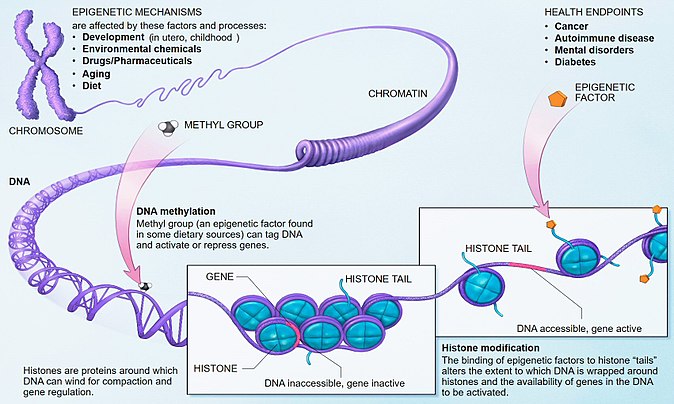
Fig. 1. Epigenetic mechanisms. Photo courtesy of National Institutes of Health via Wikipedia (https://en.wikipedia.org/wiki/Epigenetics)
Epigenetic changes can be triggered by environmental stimuli, they usually arise in particular genome regions, displaying unique patterns and are by rule reversible. Two basic roles of epigenetic regulation can be distinguished: one in the major biological processes including reprogramming during early embryogenesis, development and adaptation, and the other – in disease manifestation.
Epigenetic changes arising in response to extrinsic factors
Influence of ethanol on embryonic development, expression and methylation status of the imprinted locus Igf2/H19 in Mus musculus
Epigenetic regulation of DNA is a dynamic process that allows the information encoded in DNA to be realized depending on the organism’s needs and the changing environment. Various environmental factors can alter the epigenetic mechanisms, including substances commonly used in our society such as nicotine, caffeine, alcohol and medicines. Ethanol is a classic teratogen and its consumption during pregnancy can cause a wide range of developmental abnormalities. The mechanisms underlying alcohol teratogenicity have not yet been fully elucidated.
We investigated the relationship between ethanol-induced change in the growth of blastocysts, embryos and placentas and the level of expression and methylation status of imprinted locus Igf2/H19, after in vitro and in vivo ethanol treatment during the preimplantation period of mice pregnancy. The results showed that ethanol activates and inhibits the development of the blastocyst in an in vitro medium in a dose-dependent manner. Administered in vivo, ethanol exhibits an inhibitory dose-dependent effect on normal growth and the development of embryos and their placentas (Fig. 2). In addition, the ethanol significantly increased the fetal resorption levels in treated groups and leads to teratogenic effects.
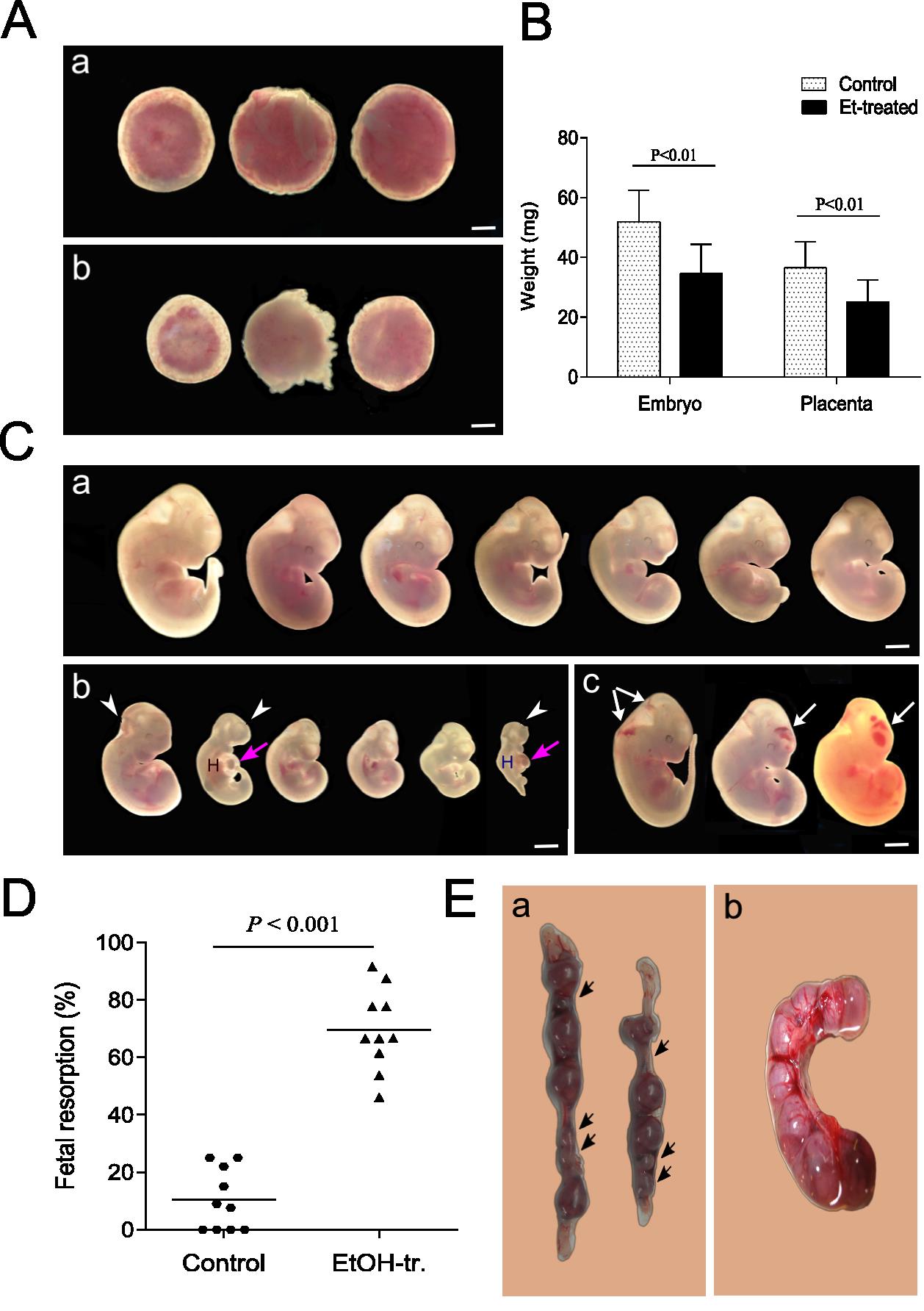
Fig. 2. Ethanol induced embryonic and placental growth retardation and increased fetal resorption. Pregnant mice were treated with 5,8 g/kg ethanol on 3.5 dpc of gestation. The analyses were performed at 11.5 dpc. (A) Normal placentas view (a), and placentas after treatment with 5,8 g/kg ethanol (b); (B) Mean embryonic and placental weight of the control, untreated mouse and after treatment with 5,8 g/kg ethanol; (C) Normal embryos view (a); embryos after treatment with 5,8 g/kg ethanol – with lower weight and developmental abnormalities (b); embryos with internal hemorrhages (white arrows) (c); (D) Fetal resorption; (E) Representative photos of uteri of alcohol treated mouse with 5,8 g/kg ethanol (a) and uterus from control, untreated mouse (b) examined on 11.5 dpc. The black arrows show the places of embryonal resorption.
Applied in vivo and in vitro along preimplantation period, ethanol changed the Igf2/H19 expression levels (Fig. 3).
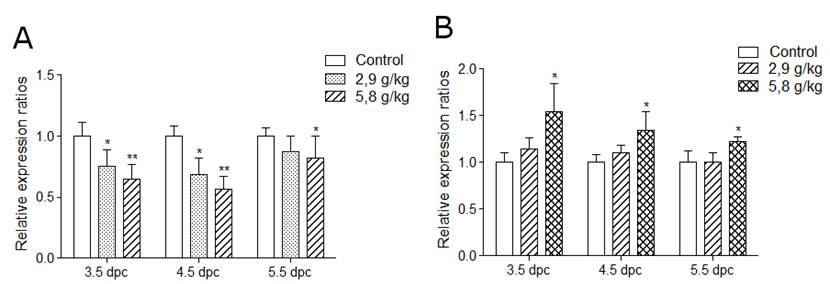
Fig. 3. Effect of 2,9 and 5,8 g/kg ethanol on embryos (A) and placentas (B) after in vivo treatment at 3.5, 4.5, and 5.5 dpc. Changes in the H19 expression were assessed at 11.5 dpc. Data is represented as means ± SEM, *p<0.05, **p<0.01.
At a treatment dose of 5.8 g/kg, ethanol leads to an alteration in the expression levels of DNA methyltransferases Dnmt1, Dnmt3b, and Dnmt3a (Fig. 4) as well as to changes in the DMRs methylation levels of Igf2/H19 locus (Fig. 5). In embryos, ethanol increased the DMRH19 methylation levels, while a decrease in the methylation of DMR0 and DMRH19 was reported in placentas.
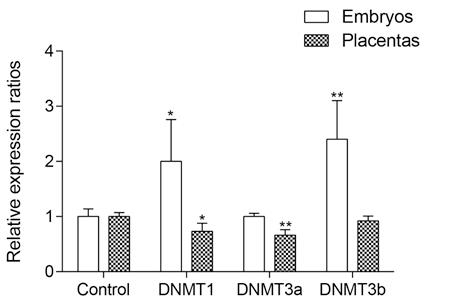
Fig. 4. Effect of 5,8 g/kg ethanol on еxpression of methyltransferases DNMT1, DNMT3a, and DNMT3b after administration at 3.5 dpc. Changes in expression levels were assessed at 11.5 dpc. Data is represented as means ± SEM, *p<0.05, **p<0.01.
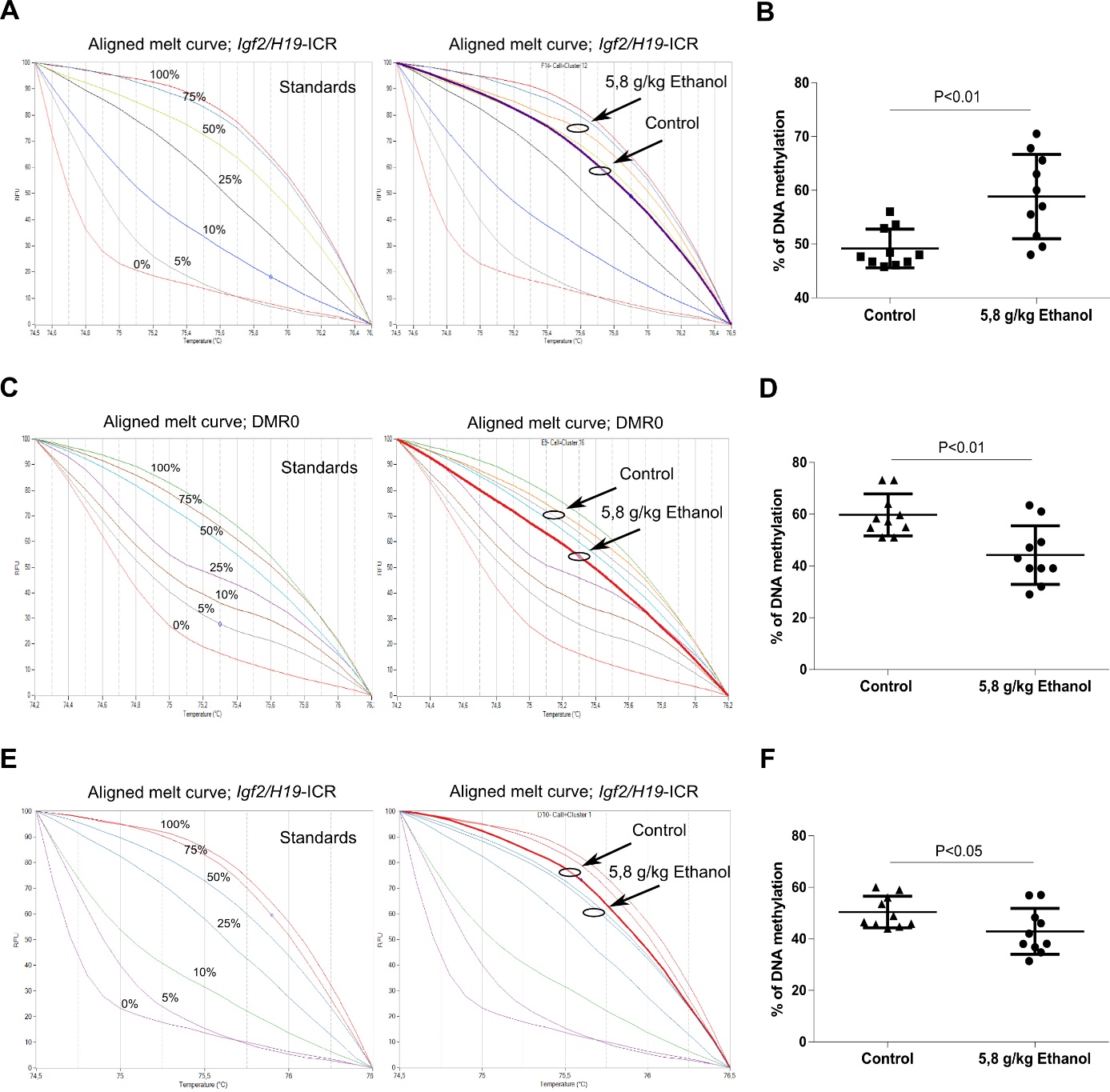
Fig. 5. Representative examples of MS-HRM and average methylation level of Igf2/H19-ICR and DMR0 in embryos and placentas. MS-HRM was used to determined change in DNA methylation after alcohol treatment based on sequence-dependent melting behavior of the amplicons. A mix of fully methylated and unmethylated DNA in a different percent give distinct methylated and unmethylated peaks in a range of 0-100% (100%, 75% 50%, 25%, 10%, 5%, and 0% methylated), which are used for standards. Aligned melt curves of standards and standards with included ethanol-treated samples and control are represented for Igf2/H19-ICR in embryos (A), DMR0 in placentas (C), Igf2/H19-ICR in placentas (E); Scatter plots showing the mean percentage methylation level of Igf2/H19-ICR in embryos (B), DMR0 in placentas (D), Igf2/H19-ICR in placentas (F) for individual samples, including ethanol-treated and controls as analyzed by MS-HRM analysis (two-tailed t-test).
The decrease of Igf2/H19– methylation of DMR0 in Igf2/H19 locus leads to alcohol-induced reduction in the placental weight. This mechanism mediates the reduction of fetal weight respectively (Fig. 6).
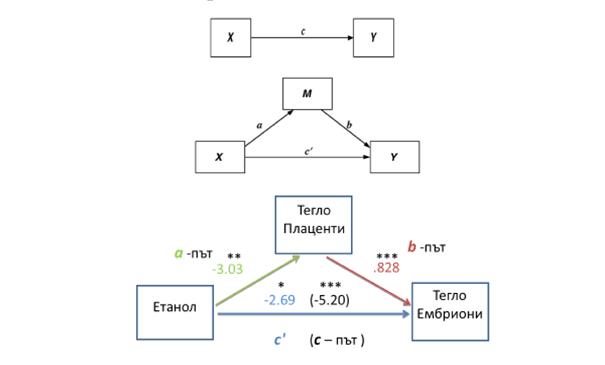
Fig. 6. Illustration of simple mediation model. Multiple regression analysis and bootstrapping algorithms were applied, using the package PROCESS and IBM SPSS Statistics 21, to assess each component of the proposed mediation model. The results from the analysis confirm a mediatory role for the placental weight in the interaction between ethanol and embryonal weight (РМ=.482, СI=-5.73 to -1.02).
In conclusion, the results show that the epigentic status and the functionality of imprinted genes are very sensitive to the effects of ethanol. Moreover, the period of implantation is highly vulnerable and susceptible to the influence of teratogenic factors.
Epigenetic changes in human diseases
Epigenetic changes were described in many complex disorders such as asthma, obesity, cardio-vascular diseases, neurodegenerative and psychological disorders, addictions, and cancer. Their unique features can explain the variability, heterogeneity and individual course of most epigenetic diseases, a phenomenon that could hardly be explained by genetic variations only.
Problematic psychoactive substances use
According to the World Health Organisation, about 2 billion people in the world use or abuse of alcohol. Regarding illegal drugs, problematic users are 27 million, or about 0,6% of the adult population. Alcohol and drug addictions, a component of more than 200 diseases, lead to serious consequences on the development, physical and psychological health of the users themselves, but also affect the well-being of the surrounding people.
A major objective of our studies is to perform a complex analysis in users of psychoactive substances taking into account and correlating psycho-social and epigenetic parameters, such as the quality of life, screening for traumatic events in person’s history, assessment of the level of expression of selected target genes and evaluation of DNA methylation patterns.
Three subject groups are being investigated: 1) Users who enter programs for opium agonists treatment; 2) Users in detoxification and psychosocial rehabilitation programs, and 3) Healthy controls. Studies follow the laboratory procedure (Fig. 7):
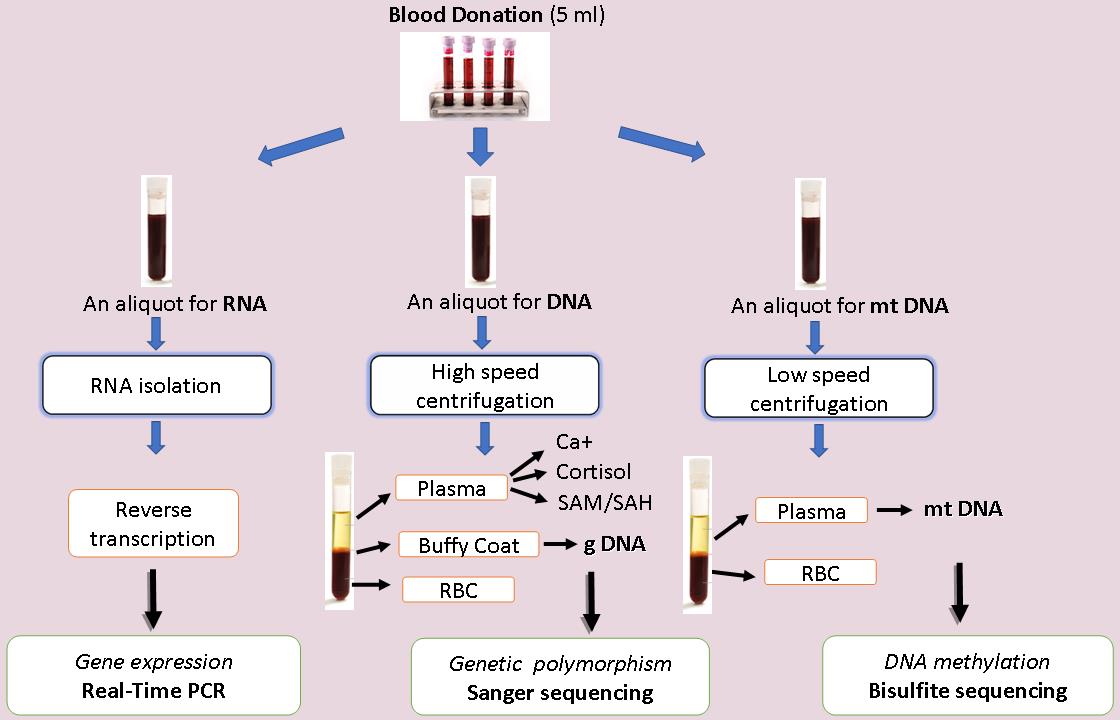
Fig. 7. Workflow of experiments.
To evaluate the level of gene expression, quantitative reverse transcription polymerase chain reaction (qRT-PCR) is applied. The following groups of target genes are being studied (Fig. 8):
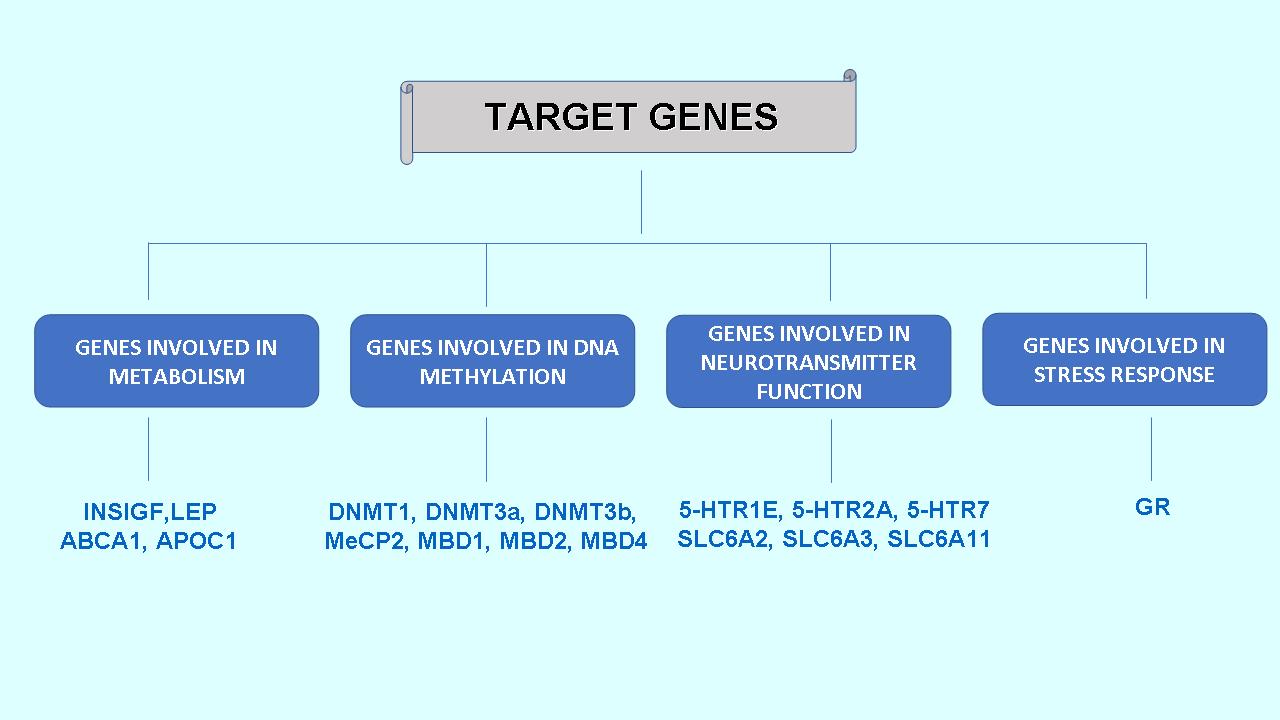
Fig. 8. Target genes analyzed in experiments.
The results will enrich the available scientific data on the pathogenic basis and etiology of psychoactive substances addictions. Deciphering the mechanisms of alcohol and drug problematic use will contribute to more accurate clinical assessment and more successful therapeutic approaches.
